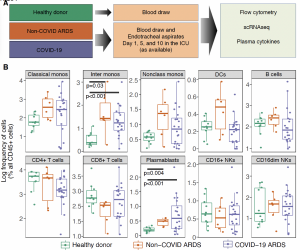
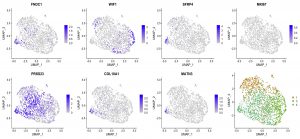
boldface: members of Benos’ lab; *corresponding author; †equal contirbution.
| “Causal discovery in high-dimensional, multicollinear datasets” | |
|
[Abstract] [Article]
|
M. Jia†, D. Yuan, T.C. Lovelace, M. Hu, P.V. Benos Front Epidemiol (2022) accepted. |
| “Essential Regression: A generalizable framework for inferring causal latent factors from multi-omic datasets” | |
| X. Bing† T. Lovelace†. F. Bunea, M. Wegkamp, S.P. Kasturi, H. Singh*, P.V. Benos*, J. Das* Patterns (2022) 3:100473. |
|
| “Critically ill COVID-19 patients 1 exhibit peripheral immune profiles predictive of mortality and reflective of SARS-CoV-2 viral burden in the lung” | |
| A.R. Cillo, A. Somasundaram, F. Shan, C. Cardello, C.J. Workman, G.D. Kitsios, A. Ruffin, S. Kunning, C. Lampenfeld, S. Onkar, S. Grebinoski, G. Deshmukh, B. Methe, C. Liu, S. Nambulli, L. Andrews, W.P. Duprex, A.V. Joglekar, P.V. Benos, P. Ray, A. Ray, B.J. McVerry, Y. Zhang, J.S. Lee, J. Das, H. Singh, A. Morris, T.C. Bruno, D.A.A. Vignali Cell Reports Med (2021) 2:100476. |
|
| “Myofibroblast transcriptome indicates SFRP2+ fibroblast progenitors in systemic sclerosis skin” | |
| T. Tabib, M. Huang, C. Morse, A. Papazoglou, R. Behera, M. Jia, M. Bulik, D. Monier, P.V. Benos, W. Chen, R. Domsic. R. Lafyatis Nature Commun (2021) 12:4384. |
|
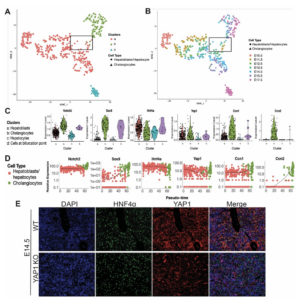
| “Compensatory hepatic adaptation accompanies permanent absence of intrahepatic biliary network due to YAP1 loss in liver progenitors” | |
| L.M. Molina, J. Zhu, Q. Li, T. Pradhan-Sundd, K. Sayed, N. Jenkins, R. Vats, S. Ko, S. Hu, M. Poddar, S.Singh, J. Tao, P. Sundd, A Singhi, S. Watkins, X. Ma, P.V. Benos, A. Feranchak, K. Nejak-Bowen, A. Watson, A. Bell, S.P. Monga Cell Reports (2021) 36:109310. |
|
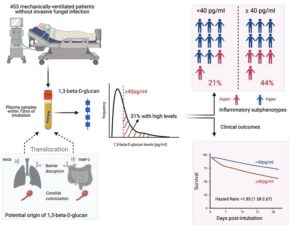
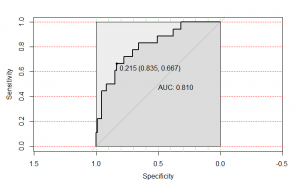
| “Plasma 1,3-beta-D-glucan levels predict adverse clinical outcomes in critical illness” | |
| G.D. Kitsios, D. Kotok, H. Yang, M.A. Finkelman, Y. Zhang, N. Britton, X. Li, M.S. Levochkina, A.K. Wagner, C. Schaefer, J.J. Villandre, R. Guo, J.W. Evankovich, W. Bain, F. Shah, Y. Zhang, B.A. Methé, P.V. Benos, B.J. McVerry, A. Morris JCI Insight (2021) 6:141277. [medRxiv] |
|
| “Transcriptomics of Bronchoalveolar Lavage Cells Identifies New Molecular Endotypes of Sarcoidosis” | |
| M. Vukmirovic, X. Yan, K.F. Gibson, M. Gulati, J.C. Schupp, G. DeIuliis, T.S. Adam, B. Hu, A. Mihaljinec, T. Woolard, H. Lynn, N. Emeagwali, E.L. Herzog, E.S. Chen, A.M. Morris, J.K. Leader, Y. Zhang, J.G.N. Garcia, L.A. Maier, R. Colman, W.P. Drake, M. Becich, H. Hochheiser, S.R. Wisniewski, P.V. Benos, D.R. Moller, A. Prasse, L.L. Koth, N. Kaminski Eur Resp J (2021) 58:2002950. [medRxiv] |
|
| “Neurological Complications Acquired during Pediatric Critical Illness: Exploratory ‘Mixed Graphical Modeling’ Analysis using Serum Biomarker Levels” | |
| V.K. Raghu, C.M. Horvat, P.M. Kochanek, E.L. Fink, R.S.B. Clark, P.V. Benos*, A. Au* Pediatric Crit Care Med (2021) 22:906-914. |
|
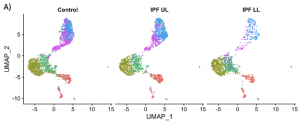
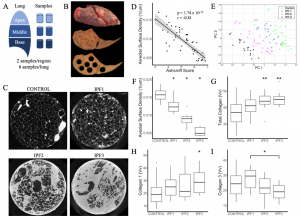
| “Reduced Proportion and Activity of Natural Killer Cells in the Lung of Patients with Idiopathic Pulmonary Fibrosis” | |
| T. Cruz†*, M. Jia†, J.C. Sembrat, T. Tabib, D.A.A. Vignali, P. Sanchez, R. Lafyatis, A.L. Mora, P.V. Benos*, M. Rojas* Am J Resp Crit Care Med (2021) 204:608-610. |
|
| “Topographic Heterogeneity of Lung Microbiota in End-Stage Idiopathic Pulmonary Fibrosis: The Microbiome in Lung Explants-2 (MiLEs-2) Study” | |
| E. Valenzi, H. Yang, J.C. Sembrat, L. Yang, S. Winters, R. Nettles, D.J. Kass, S. Qin, X. Wang, M. Myerburg, B. Methe, A. Fitch, J. Alder, P.V. Benos, B.J. McVerry, M. Rojas, A. Morris, G.D. Kitsios Thorax (2021) 76:239-247. [medRxiv] |
|
| “Human ex vivo lung perfusion: A novel model to study human lung diseases” | |
| N. Cárdenes†, J. Sembrat†, K. Noda†, T. Lovelace†, D. Álvarez, H.E. Trejo Bittar, B.J. Philips, M. Nouraie, P.V. Benos,, P.G. Sánchez, M. Rojas Scientific Reports (2021) 11:490. |
|
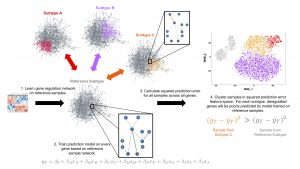
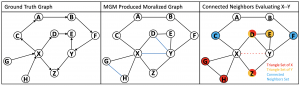
| “Improving Constraint-Based Causal Discovery from Moralized Graphs” | |
 [Abstract] [Article] [Supplementary]
|
A. Fuccello, D. Yuan, P.V. Benos, V.K. Raghu NeurIPS 2020 workshop on Causality (2020) accepted. Presented: 11-Dec-2020. |
| “miR-1207-5p can contribute to dysregulation of inflammatory response in COVID-19 via targeting SARS-CoV-2 RNA” | |
| G. Bertolazzi, C. Cipollina, P.V. Benos, M. Tumminello, C. Coronnello Frontiers in Cellular and Infection Microbiology (2020) 10:586592. |
|
| “Interpretable Factors in scRNA-seq Data with Disentangled Generative Models” | |
|
[Abstract] [Article]
|
H. Mao, , M.J. Broerman, P.V. Benos, 2020 IEEE BIBE International Conference (2020) accepted. Presented: 26-Oct-2020. |
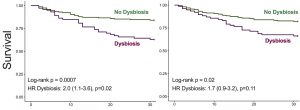
| “Respiratory Tract Dysbiosis is Associated With Worse Outcomes in Mechanically-Ventilated Patients” | |
| G.D. Kitsios, H. Yang, , L. Yang, S. Qin, A. Fitch, X.-H. Wang, K. Fair, J. Evankovich, W. Bain, F Shah, K. Li, B. Methé, P.V. Benos, A. Morris, B.J. McVerry, Am J Resp Crit Care Med (2020) 202:1666-1677. |
|
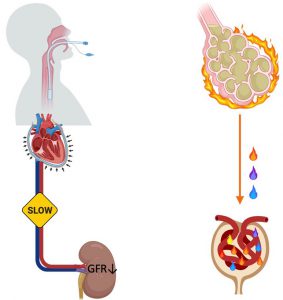
| “Protecting the lungs but hurting the kidneys: causal inference study for the risk of ventilation-induced kidney injury in ARDS” | |
| H. Yang, P.V. Benos, G.D. Kitsios, Annals in Transl Med (2020) 8:985. [Editorial] |
|
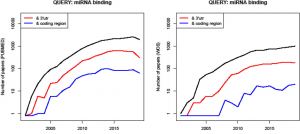
| “An improvement of ComiR algorithm for microRNA target prediction by exploiting coding region sequences of mRNAs” | |
| G. Bertolazzi, P.V. Benos, M. Tumminello, C. Coronnello, BMC Bioinformatics (2020) 21(Suppl 8):363. |
|
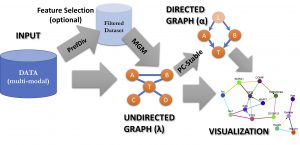
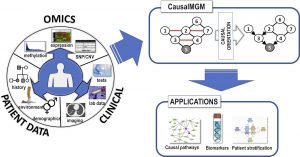
| “CausalMGM: An interactive web-based causal discovery tool” | |
| X. Ge†, V.K. Raghu†, P.K. Chrysanthis, P.V. Benos, Nucl Acids Res (2020) 48 (Web Server Issue):W597–W602. |
|
| “The evolution of radiographic edema in ARDS and its association with clinical outcomes: a prospective cohort study in adult patients” | |
| D. Kotok, J.W. Evankovich, W. Bain, D.G. Dunlap, F. Shah, L. Yang, Y. Zhang, D.V. Manatakis, P.V. Benos, I.J. Barbash, S.F. Rapport, J.S. Lee, A. Morris, B.J. McVerry, G.D. Kitsios, J Crit Care (2020) 56:222-228. |
|
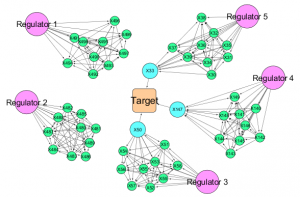
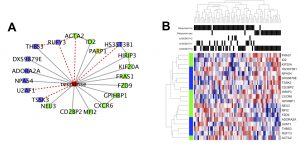
| “Causal network perturbations for instance-specific analysis of single cell and disease samples” | |
| K.L. Buschur, M. Chikina, P.V. Benos Bioinformatics (2020) 36:2515–2521. |
|
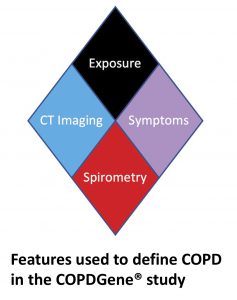
| “COPDGene 2019: Redefining the Diagnosis of Chronic Obstructive Pulmonary Disease” | |
| K.E. Lowe, E.A. Regan, A. Anzueto, E. Austin, J.H.M. Austin, Beaty TH, P.V. Benos, …, E.K. Silverman, J.D. Crapo Chronic Obstructive Pulm Dis (2019) 6:384-399. |
|
| “Host-Response Subphenotypes Offer Prognostic Enrichment in Patients With or at Risk for Acute Respiratory Distress Syndrome” | |
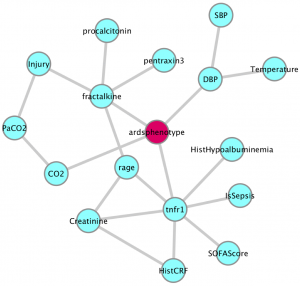 |
G.D. Kitsios, L. Yang, D.V. Manatakis, M. Nouraie, J. Evankovich, W. Bain, D.D. Dunlap, F. Shah, I.J Barbash, S.F. Rapport, Y. Zhang, R.S. DeSensi, N.M. Weathington, B.B. Chen, P. Ray, R.K. Mallampalli, P.V. Benos, J.S. Lee, A. Morris, B.J. McVerry Critical Care Medicine (2019) 47:1724-1734. |
| “A dynamic regulatory model of gene expression changes in differentially affected regions of the human IPF lung” | |
| J.E. McDonough, F. Ahangari1, Q. Li, S. Jain, S.E. Verleden, J. Herazo-Maya, M. Vukmirovic, G. Deluliis, A. Tzouvelekis, N. Tanabe, F. Chu, X. Yan, J. Verschakelen, R.J. Homer, D.V. Manatakis, J. Zhang, J. Ding, K. Maes, L. De Sadeleer, R. Vos, A. Neyrinck, P.V. Benos, Z. Bar-Joseph, D. Tantin, J.C. Hogg, B.M. Vanaudenaerde, W.A. Wuyts, N. Kaminski JCI Insight (2019) 4:e131597. |
|
| “Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis” | |
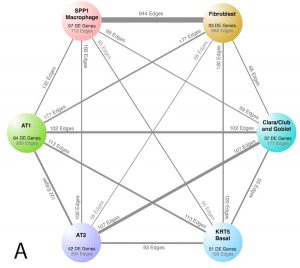 |
C. Morse, T. Tabib, J. Sembrat, K.L. Buschur, H.T. Bittar, E. Valenzi, Y. Jiang, D.J. Kass, K. Gibson, W. Chen, A. Mora, P.V. Benos, M. Rojas, R. Lafyatis European Respiratory Journal (2019) 54:1802441. |
| “A pipeline for integrated theory and data-driven modeling of genomic and clinical data” | |
|
[Abstract] [Article]
|
V.K. Raghu, X. Ge, A. Balajee, D. Shirer, I. Das, P.V. Benos, P.K. Chrysanthis BioKDD2019 (2019) accepted. |
| “Expression patterns of small numbers of transcripts from functionally-related pathways predict survival in multiple cancers” | |
| J. Mandel, H. Wang, D.P. Normolle, W. Chen, Q. Yan, P.C. Lucas, P.V. Benos, E.V. Prochownik BMC Cancer (2019) 19:686. |
|
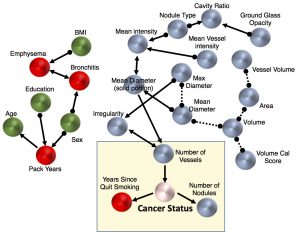
| “Feasibility of lung cancer prediction from low-dose CT scan and smoking factors using causal models” | |
| V.K. Raghu, W. Zhao, J. Pu, J.K. Leader, R. Wang, J. Herman, J.-M. Yuan, P.V. Benos*, D.O. Wilson Thorax (2019) 74:643-649. |
|
| “PARP1 rs1805407 Increases Sensitivity to PARP1 Inhibitors in Cancer Cells Suggesting an Improved Therapeutic Strategy” | |
| I. Abecassis†, A.J. Sedgewick†, M. Romkes, S. Buch, T. Nukui, M.G. Kapetanaki, A. Vogt, J.M. Kirkwood, P.V. Benos*, H. Tawbi* Scientific Reports (2019) 9:3309. |
|
| “Mixed Graphical Models for Integrative Causal Analysis with Application to Chronic Lung Disease Diagnosis and Prognosis” | |
| A.J. Sedgewick, K. Buschur, I. Shi, J.D. Ramsey, V.K. Raghu, D.V. Manatakis, Y. Zhang, J. Bon, D. Chandra, C. Karoleski, F.C. Sciurba, P. Spirtes, C. Glymour, P.V. Benos Bioinformatics (2019) 35:1204-1212. |
|
| “Mitochondria are a substrate of cellular memory” | |
| A. Cheikhi, C. Wallace, C.S. Croix, C. Cohen, W.Y. Tang, P. Wipf, P.V. Benos, F. Ambrosio, A. Barchowsky Free Radic Biol Med. (2019) 130:528-541. |
|