|
|
|
|
PDB Code |
n |
Resolution (Å) |
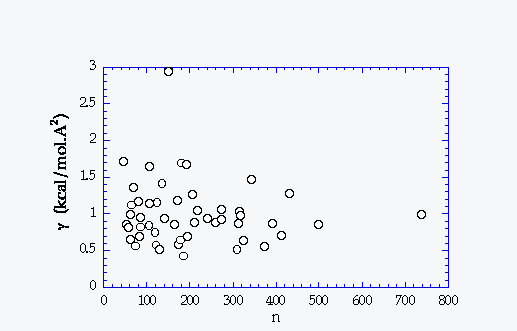
g (kcal/mol.Å 2) |
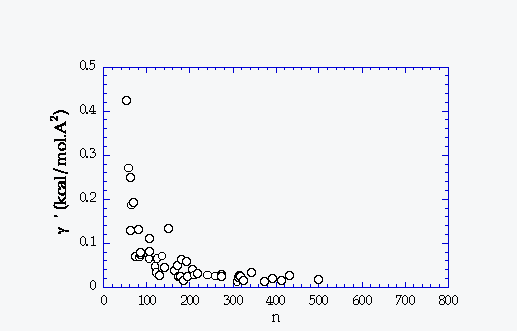
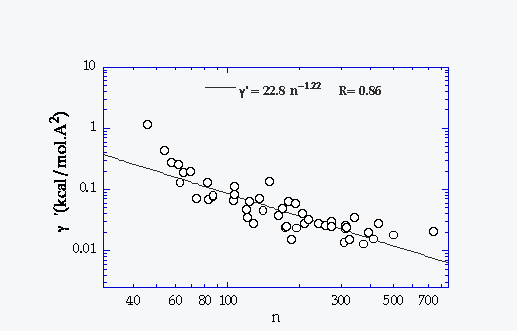
g Resolution |
|
1crn |
46 |
1.5 |
1.71 |
1.14 |
|
4rxn |
54 |
2 |
0.847 |
0.423 |
|
5pti |
58 |
1.8 |
0.810 |
0.270 |
|
5ebx |
62 |
2 |
0.995 |
0.249 |
|
1r69 |
63 |
2 |
0.647 |
0.129 |
|
1sn3 |
65 |
1.8 |
1.12 |
0.187 |
|
2utg |
70 |
1.64 |
1.35 |
0.193 |
|
1hoe |
74 |
2 |
0.559 |
0.0699 |
|
451c |
82 |
1.6 |
1.17 |
0.130 |
|
2ci2 |
83 |
2 |
0.683 |
0.0683 |
|
2gn5 |
87 |
2.3 |
0.817 |
0.0743 |
|
1aba |
8 |
1.45 |
0.946 |
0.0789 |
|
256b |
106 |
1.4 |
0.845 |
0.0650 |
|
1fkf |
107 |
1.7 |
1.13 |
0.0809 |
|
2cdv |
107 |
1.8 |
1.65 |
0.110 |
|
1paz |
120 |
1.55 |
0.743 |
0.0465 |
|
1alc |
122 |
1.7 |
0.573 |
0.0337 |
|
9rsa |
124 |
1.8 |
1.15 |
0.0637 |
|
2aza |
129 |
1.8 |
0.513 |
0.0270 |
|
1eca |
136 |
1.4 |
1.41 |
0.0705 |
|
1hds |
141 |
1.98 |
0.929 |
0.0442 |
|
2sod |
151 |
2.0 |
2.93 |
0.133 |
|
3lzm |
164 |
1.7 |
0.848 |
0.0369 |
|
9wga |
171 |
1.8 |
1.17 |
0.0489 |
|
1rbp |
175 |
2.0 |
0.583 |
0.0233 |
|
1cc5 |
178 |
2.5 |
0.642 |
0.0247 |
|
2ltn |
181 |
1.7 |
1.69 |
0.0626 |
|
8dfr |
186 |
1.7 |
0.424 |
0.0151 |
|
3adk |
194 |
2.1 |
1.67 |
0.0577 |
|
2stv |
195 |
2.5 |
0.695 |
0.0232 |
|
3gap |
208 |
2.5 |
1.26 |
0.0405 |
|
9pap |
212 |
1.65 |
0.883 |
0.0276 |
|
3cla |
219 |
1.75 |
1.05 |
0.0317 |
|
3sgb |
242 |
1.8 |
0.934 |
0.0275 |
|
2cab |
260 |
2.0 |
0.879 |
0.0251 |
|
2sec |
275 |
1.8 |
1.05 |
0.0293 |
|
2sni |
275. |
2.1 |
0.924 |
0.0250 |
|
8atc |
310 |
2.5 |
0.508 |
0.0134 |
|
1fnr |
314 |
2.2 |
0.866 |
0.0222 |
|
6tmn |
316 |
1.6 |
1.03 |
0.0257 |
|
2ts1 |
319 |
2.3 |
0.975 |
0.0238 |
|
4pep |
326 |
1.8 |
0.638 |
0.0152 |
|
2liv |
344 |
2.4 |
1.47 |
0.0342 |
|
8adh |
374 |
2.4 |
0.555 |
0.0126 |
|
1wsy |
393 |
2.5 |
0.867 |
0.0193 |
|
2ccp |
414 |
2.2 |
0.702 |
0.0153 |
|
5cts |
433 |
1.9 |
1.27 |
0.0271 |
|
8cat |
500 |
2.5 |
0.854 |
0.0178 |
|
1cdk |
740 |
2.0 |
0.988 |
0.0200 |